Section: New Results
Development of RDKit Smiles Manager SAMSON Elements
Participant : Yassine Naimi.
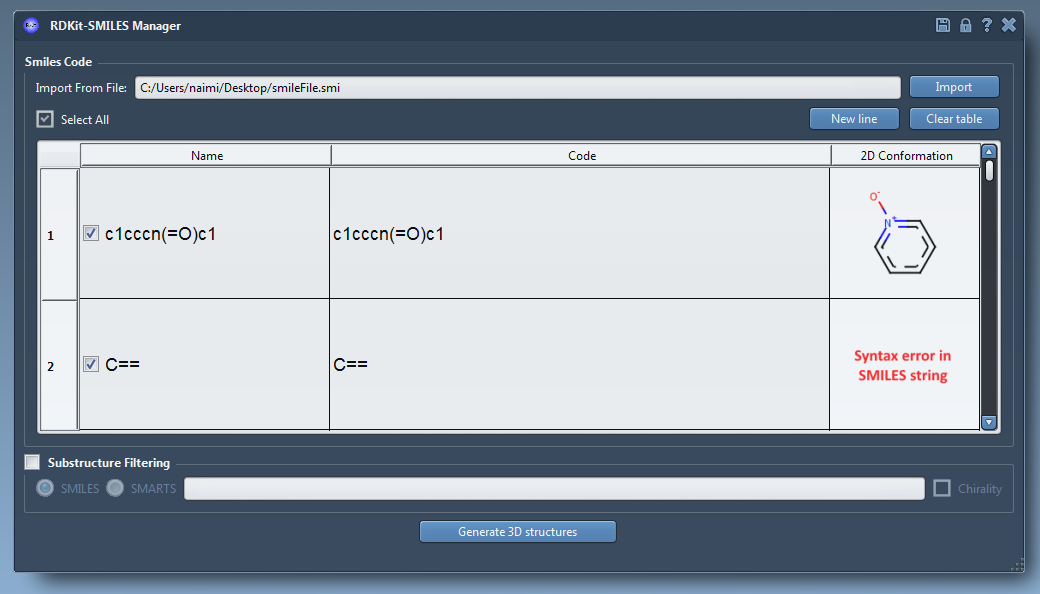
We integrated RDKit, an open-source collection of cheminformatics and machine-learning software written in C++ and Python, in SAMSON. One of RDKit's features is the conversion of molecules from their SMILES code to a 2D and 3D structures. Therefore, it is now possible to use these features in the SAMSON platform. SMILES code files (.smi) or text files (.txt) containing several SMILES codes can be read using the import button (19).
Users can manage the imported data with several manners (modify the SMILES code, add manually a new SMILES code, assign names to the molecules…). Also, 2D depictions of the SMILES code are generated on the fly (20). When a SMILES code is invalid an error image is automatically generated. By right-clicking on these images, users can open or generate the 3D structure in SAMSON or save the image as png or svg. The main feature of this element is to generate 3D structures from imported/written SMILES codes. After selecting the molecules, users can click on the Generate 3D structures button. Few seconds later, the 3D structures of the molecules (presenting a valid SMILES code) are added to the SAMSON document view. Finally, RDKit provides a feature to filter the selected molecules using a substructure pattern (SMILES or SMARTS). By default in RDKit, information about stereochemistry is not used in substructure searches but this can be changed by using the chirality. For information, the name of the molecules that did not include the given pattern are displayed in a pop-up.